

📍 454 pyrosequencing: These folks take advantage of clonal amplification through emulsion PCR. 📍 Illumina: They use single-stranded DNA-binding proteins to amplify DNA and then add fluorescent-labeled deoxynucleoside triphosphates to bridge the amplified DNA template. 💪Here are some of the short-read sequencing technologies making waves in the scientific community:
Short reads are effective for applications aimed at counting the abundance of specific sequences, identifying variants within otherwise well-conserved sequences, or profiling the expression of particular transcripts. Compared to traditional methods, it's faster and more cost-effective.🎯 It's also interesting to note that the "original" library molecules serve only to make the first copy of a surface bound template strand, and are then likely washed away.Short-read sequencing has revolutionized the field of biomedical research. The things to remember are that the glass surface contains two primers, and each library molecule contains complementary ends to each of the surface primers.
BRIDGE PCR VS EMULSION PCR PATCH
The end result is a little patch of molecules, like a little tree stand, that all began with a single template, that got copied by "bridging" of the template between the surface bound forward and reverse PCR primers. During each PCR cycle, these new templates, bend over and form "bridges" to their complementary PCR primers, which are all over the surface. That matching primer then gets extended, forming yet another template.
BRIDGE PCR VS EMULSION PCR FREE
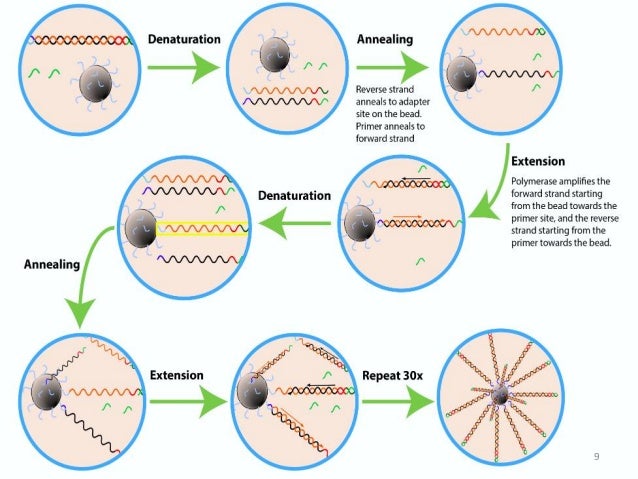
During the second PCR cycle, this free end, which has a sequence complimentary to the other of the two surface primers, bends over and forms a "bridge" to a matching primer, serving as a template. The second end is free to wave around like a blade of grass. Since the new template is stuck to the surface, it can't move. But when the first cyle of PCR happens, the matching primer - which is covalently bound to the surface, gets extended and converted into a new template strand. The template strand is not actually covalently bound to the surface, it simply interacts with one of the surface primers through base pairing. This second strand also contains both primer sequences on each end (just as the original template molecule did). Thus in a PCR reaction, the 5' end of any given template will match a surface primer, and that primer will get extended across the template to make a second strand. Each end of every library molecule matches one of the two primers on the glass surface. The library is diluted such that molecules spread out across the surface at some optimum density - giving each one enough space to "build it's own house" as it were. This is what is done for Illumina sequencing. Wherever a template molecule lands, it is within reach of the forward and reverse primers bound to the surface. an Illumina Library), and pipette them in a solution over the glass surface. Then, when you want to do PCR, take your template molecules (e.g. You could instead take a mixture of the two oligos and spread them out over a glass surface, allowing them to become covalently attached to the surface (so they never come off the surface). However, there's no reason why they have to be in solution. Normally these oligos are free in solution in a tube, and with every PCR cycle, they find a match in a template sequence and are extended to create a new template strand. PCR is typically performed with two different oligos, often referred to as forward and reverse primers, that match some target template sequence. This process occurs in what is referred to as Illumina's "cluster station", an automated flow cell processor. Repeated denaturation and extension results in localized amplification of single molecules in millions of unique locations across the flow cell surface. Priming occurs as the free/distal end of a ligated fragment "bridges" to a complementary oligo on the surface. Single-stranded, adapter-ligated fragments are bound to the surface of the flow cell exposed to reagents for polyermase-based extension. The flow cell surface is coated with single stranded oligonucleotides that correspond to the sequences of the adapters ligated during the sample preparation stage. In contrast to the 454 and ABI methods which use a bead-based emulsion PCR to generate "polonies", Illumina utilizes a unique "bridged" amplification reaction that occurs on the surface of the flow cell.


 0 kommentar(er)
0 kommentar(er)
